Please note that a variety of docking management applications exist to assist you in this task. Vina avoids imposing artificial restrictions, such as the number of atoms in the input, the number of torsions, the size of the search space, the exhaustiveness of the search, etc. Why don't the results change when I change the partial charges? Why do I not get the correct bound conformation? You are using a 2D flat ligand as input. You might be able to do that, but AutoDock Vina is designed only for receptor-ligand docking. Will you answer my questions about Vina if I email or call you?
| Uploader: | Faerisar |
| Date Added: | 5 March 2013 |
| File Size: | 22.68 Mb |
| Operating Systems: | Windows NT/2000/XP/2003/2003/7/8/10 MacOS 10/X |
| Downloads: | 1701 |
| Price: | Free* [*Free Regsitration Required] |
See Features It should be noted that the predictive accuracy varies a lot depending on the target, so it makes sense to evaluate AutoDock Vina against your particular target first, if you have known actives, or a bound native ligand structure, before ordering compounds. AFAIK, AutoDock4 used grid too, but the grid has to be calculated in advance, it also uses 46 configuration file and zutodock configuration file I see incorrectly oriented hydrogens in the ligand and flexible receptor parts.
If you are not doing redocking, i. It also uses the same type of structure format PDBQT for maximum compatibility with auxiliary software. Current status and future challenges". Yes is really so simple!
autodock vina free download for windows 10 64 bit
While evaluating any docking engine in a retrospective virtual screen, it might make sense to select decoys of similar size, and perhaps other physical characteristics, to your known actives. AutoDock consists of two main programs: Any other way to view the results? Expert information Infrastructure Perun Scheduling system. Journal of Medicinal Chemistry.

The instructions on preparation of flexible receptor sidechains are also included. Install Boost Install Boost. The scoring function has changed since the tutorial was recorded, but only in the part that is independent of the conformation: AutoDockTools is the preferred application to visualize the grid - zutodock space, see the above mentioned tutorial.
However, please do not include requests for assistance along with your bug report. From Wikipedia, the free encyclopedia. All that is required is the structures of the molecules being docked and the specification of the search space including the binding site. The minimum of the scoring function correponds to the correct conformation, but the search algorithm has trouble finding it. Therefore, in the output, some hydrogen atoms can be expected to be positioned randomly but consistent with the covalent structure.
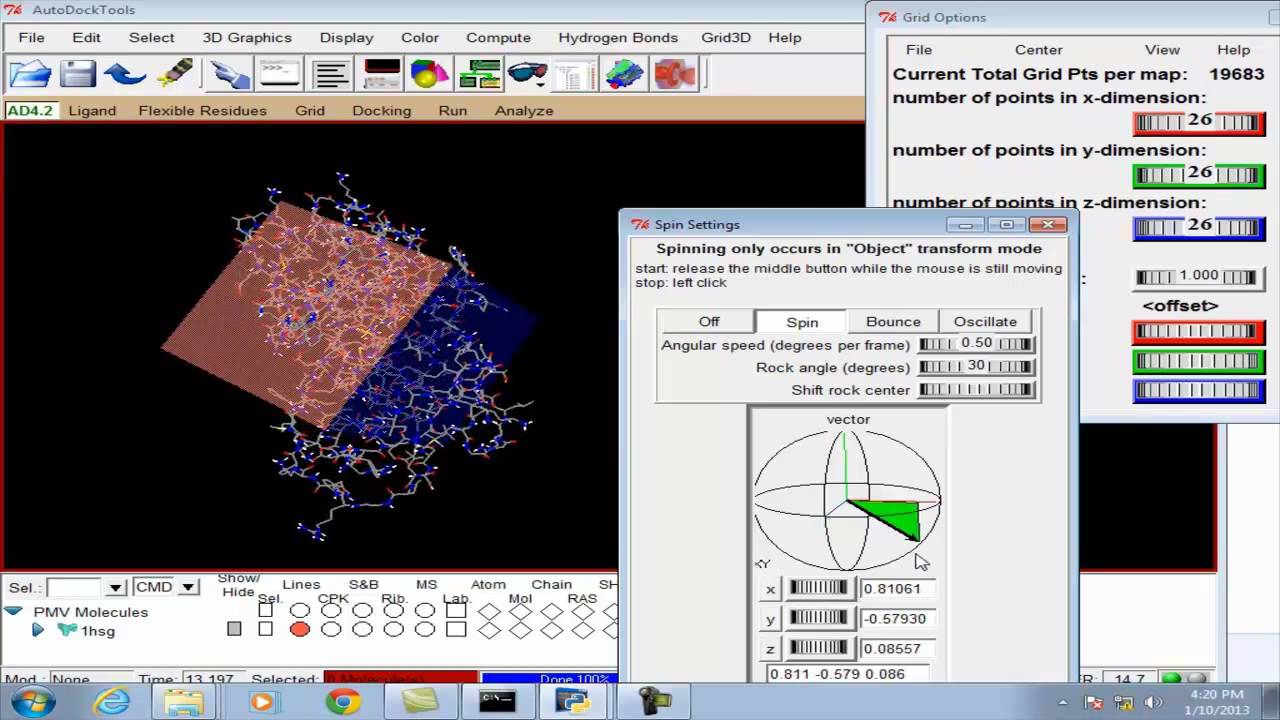
Why was it developed? Docking is an approximate approach. Why do my results look weird in PyMOL? Yes, Vina uses a grid which is defined by its center and number of points in each dimension. To display the available command line arguments type "vina --help".
Download AutoDock Vina
If you are coming from AutoDock 4, a very common mistake is to specify the search space in "points" 0. The examples below assume that Bash is your shell. PyMOL is one of the most popular programs for molecular visualization and can be used for viewing the docking results OpenBabel can be used to convert among various structure file formats, assign the protonation states, etc.
I don't have aitodock computing resources to run a virtual vuna. How do I use flexible side chains? This option specifies the maximum number of binding modes to output. For optimal performance, remember to vinx using the Release mode. This setting can be changed in the control panel or system preferences. History Brief summaries of changes between versions can be found here.
How is this possible?

No comments:
Post a Comment